Data enrichment is one of common data engineering tasks. It's relatively easy to implement with static datasets because of the data availability. However, this apparently easy task can become a nightmare if used with inappropriate technologies.
Data Engineering Design Patterns

Looking for a book that defines and solves most common data engineering problems? I wrote
one on that topic! You can read it online
on the O'Reilly platform,
or get a print copy on Amazon.
I also help solve your data engineering problems 👉 contact@waitingforcode.com 📩
The idea for this blog post came after one of my Structured Streaming workshops. One of the participants asked a question whether refreshTable API can help refresh the table used in the join with the streaming DataFrame. I don't answer it right now but you should get it by the end of the article!
Old world
To put it simply, you have two choices for joining streaming and batch DataFrames. The first one uses table file formats like Delta Lake or Apache Iceberg and it should be the preferred one for one important property. The refresh capability. Well, I'm realizing I partially answered the question and you don't need to read the whole blog post. But it doesn't matter, you'll probably stay here to know why!
An alternative is anything else than the table file formats will put you in trouble. Let's take this example where we first create a table, later start the stream job with the join, and finally overwrite the table's content. The shortened version of these three jobs is below:
// create table
val dataset = Seq((0, "A"), (1, "B"), (2, "C")).toDF("number", "letter")
dataset.write.saveAsTable("letters_enrichment_table")
// streaming job with the join
val inputDataFrame = sparkSession.readStream.format("rate").option("rowsPerSecond", 10).load().withColumn("number", $"value" % 3)
val referenceDatasetTable = sparkSession.table("letters_enrichment_table")
val joinedDataset = inputDataFrame.join(referenceDatasetTable, Seq("number"), "inner")
joinedDataset.writeStream.format("console").start().awaitTermination()
// update table
val dataset = Seq((0, "AA"), (1, "BB"), (2, "CC"), (3, "DD")).toDF("number", "letter")
dataset.write.mode("overwrite").saveAsTable("letters_enrichment_table")
The job will be running fine until you execute the last step. When it happens, the streaming job will fail because of this exception:
Caused by: java.util.concurrent.ExecutionException: org.apache.spark.SparkException: Job aborted due to stage failure: Task 0 in stage 364.0 failed 1 times, most recent failure: Lost task 0.0 in stage 364.0 (TID 1995) (192.168.1.55 executor driver): org.apache.spark.SparkFileNotFoundException: File file:/tmp/join_structured_streaming/table/warehouse/letters_enrichment_table/part-00001-be1edd0f-673d-43c9-8722-3da96d2dc555-c000.snappy.parquet does not exist It is possible the underlying files have been updated. You can explicitly invalidate the cache in Spark by running 'REFRESH TABLE tableName' command in SQL or by recreating the Dataset/DataFrame involved.
Good thing with this exception is the solution hint. So, let's try to rewrite the streaming job and refresh the table:
val inputDataFrame = sparkSession.readStream.format("rate").option("rowsPerSecond", 10).load().withColumn("number", $"value" % 3)
sparkSession.sql("REFRESH TABLE letters_enrichment_table")
val referenceDatasetTable = sparkSession.table("letters_enrichment_table")
val joinedDataset = inputDataFrame.join(referenceDatasetTable, Seq("number"), "inner")
joinedDataset.writeStream.format("console").start().awaitTermination()
Unfortunately, it still fails for the same reason. I did this mistake by putting the REFRESH before in the main flow as it's how many beginners see stream processing with Structured Streaming. They consider it as a whole job iteratively executed whereas only the parts connected to the streams are. An easy solution would be to include the join in the foreachBatch but then it behaves like join of two batch DataFrames:
inputDataFrame.writeStream.foreachBatch((rateDataFrame: DataFrame, batchNumber: Long) => {
sparkSession.sql("REFRESH TABLE letters_enrichment_table")
val referenceDatasetTable = sparkSession.table("letters_enrichment_table")
val joinedDataset = inputDataFrame.join(referenceDatasetTable, Seq("number"), "inner")
joinedDataset.show(truncate = false)
()
}).start().awaitTermination()
Well, bad luck, it doesn't work either. The error is the same and the issue is related to the overwrite mode of the updater. Since it removes the files, the error message still mentions the deleted files
Caused by: org.apache.spark.SparkFileNotFoundException: File file:/tmp/join_structured_streaming/table/warehouse/letters_enrichment_table/part-00000-a5301815-99cf-4f57-a8dd-3e63d99d7081-c000.snappy.parquet does not exist It is possible the underlying files have been updated. You can explicitly invalidate the cache in Spark by running 'REFRESH TABLE tableName' command in SQL or by recreating the Dataset/DataFrame involved.
Refresh in a listener?
It's not a good idea either. The listeners are asynchronous. Consequently, they won't execute before the next micro-batch and you can still encounter the missing files issue. I wrote about TODO: link some time ago.
But you'll see later that the root cause is not the same as for the first error. However, let me show you a modern approach.
New world
In the new world, you use a table file format that not only provides atomicity guarantees but also an automatic refresh for the joined dataset. Look at the same example but with the format changes to "delta":
// create table
val dataset = Seq((0, "A"), (1, "B"), (2, "C")).toDF("number", "letter")
dataset.write.format("delta").saveAsTable("letters_enrichment_table")
// streaming job with the join
// same as for the Old world
// update table
val dataset = Seq((0, "AA"), (1, "BB"), (2, "CC"), (3, "DD")).toDF("number", "letter")
dataset.write.format("delta").mode("overwrite").saveAsTable("letters_enrichment_table")
The result? The update not only doesn't break the stream but also provides fresh data in the next micro-batch:
------------------------------------------- Batch: 22 ------------------------------------------- +------+--------------------+-----+------+ |number| timestamp|value|letter| +------+--------------------+-----+------+ | 2|2024-01-10 08:55:...| 590| C| | 1|2024-01-10 08:55:...| 598| B| | 0|2024-01-10 08:55:...| 606| A| ... | 1|2024-01-10 08:55:...| 604| B| | 0|2024-01-10 08:55:...| 597| A| | 2|2024-01-10 08:55:...| 605| C| +------+--------------------+-----+------+ ------------------------------------------- Batch: 23 ------------------------------------------- +------+--------------------+-----+------+ |number| timestamp|value|letter| +------+--------------------+-----+------+ | 1|2024-01-10 08:55:...| 610| BB| | 0|2024-01-10 08:55:...| 618| AA| | 2|2024-01-10 08:55:...| 626| CC| | 2|2024-01-10 08:55:...| 611| CC| ... | 2|2024-01-10 08:55:...| 623| CC| | 1|2024-01-10 08:55:...| 616| BB| | 0|2024-01-10 08:55:...| 624| AA| | 2|2024-01-10 08:55:...| 617| CC| | 1|2024-01-10 08:55:...| 625| BB| +------+--------------------+-----+------+
Why?
To understand this difference, let's analyze how Apache Spark Structured Streaming sees both table overwrites:
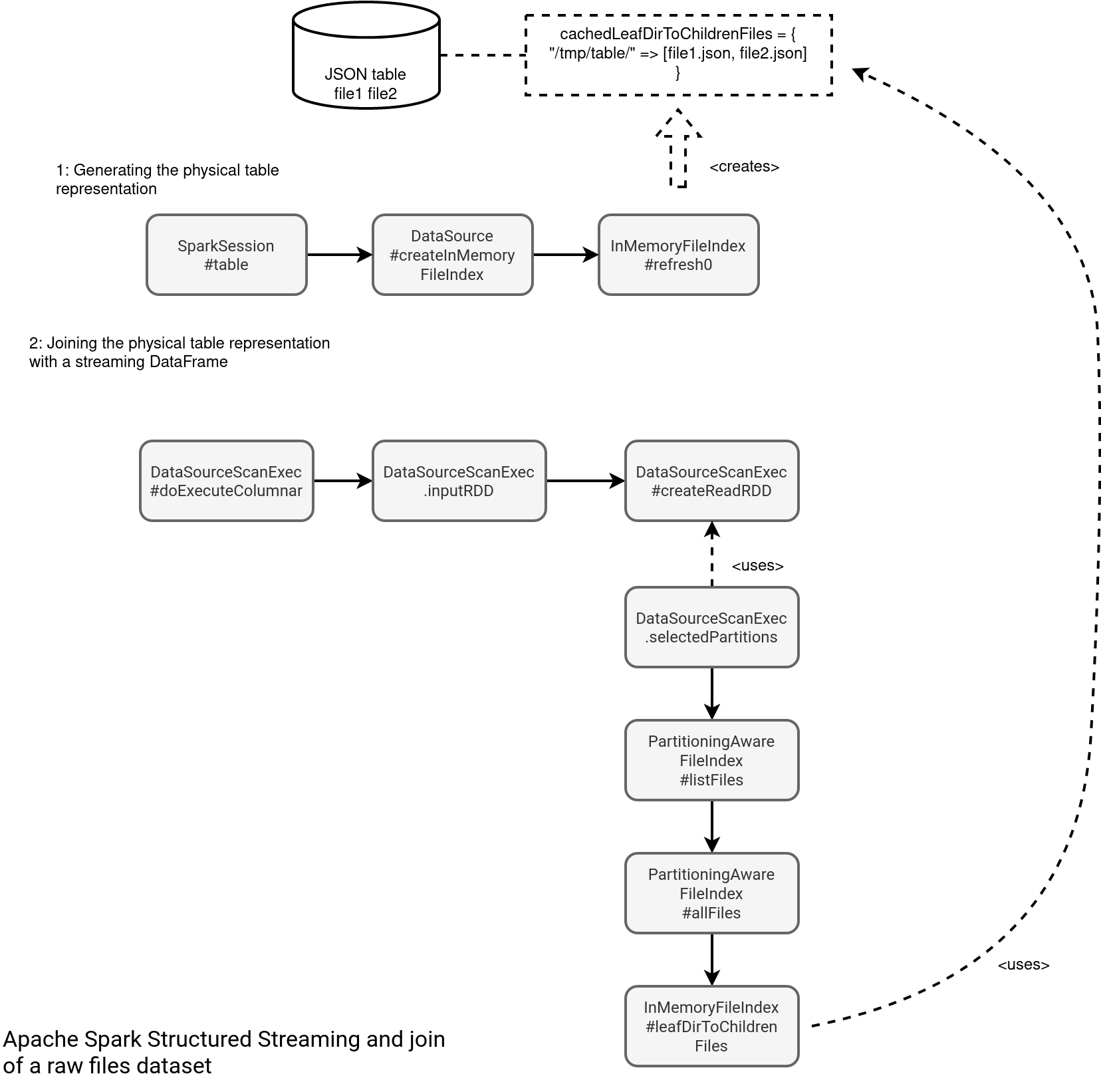
As you can see in the picture, the whole issue is with the in-memory index created at the table definition time. It caches all files representing a table to avoid often costly files listing operations. But wait a minute, if it's cached, the REFRESH command should help, shouldn't it? Well, yes and no. The RefreshTableCommand indeed refreshes the in-memory index:
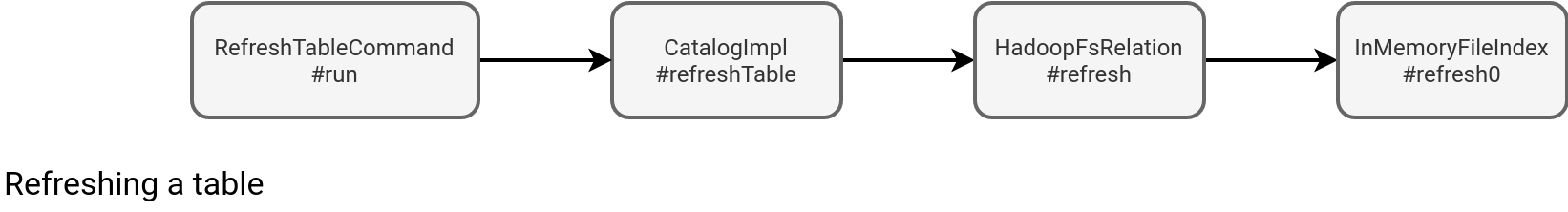
However, it doesn't guarantee the refreshed table will always synchronize with the update. If they're concurrent, you've a lot of chances to see the missing file errors. Otherwise, it can work. But still, it's more risky than the join with a table file formats dataset. Why? Because Delta Lake already provides the index with all files to read. Let's take a look at the index generation flow:
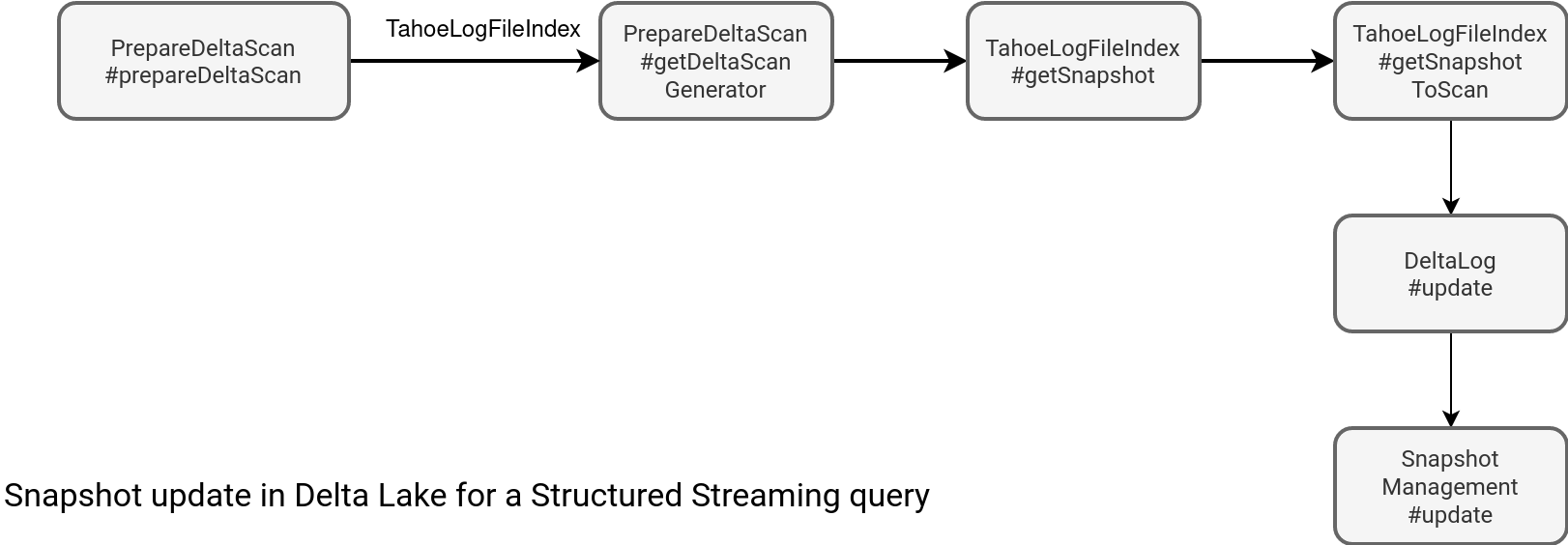
The following workflow runs at each micro-batch. It means that whenever a writer creates a new Delta table version, the snapshot update process will fetch it from the table location. How? By listing either all commit files (_delta_log location) starting from the most recent checkpoint, or from the beginning, if the checkpoint doesn't exist yet. Later, it generates a so called log segment with all valid data files. This segment also gives the information about the last table version since it corresponds to the version of the last commit file:
// SnapshotManagement#getLogSegmentForVersion(versionToLoad: Option[Long]) val lastCheckpointVersion = getCheckpointVersion(lastCheckpointInfo, oldCheckpointProviderOpt) val listingStartVersion = Math.max(0L, lastCheckpointVersion) val includeMinorCompactions = spark.conf.get(DeltaSQLConf.DELTALOG_MINOR_COMPACTION_USE_FOR_READS) val newFiles = listDeltaCompactedDeltaAndCheckpointFiles( startVersion = listingStartVersion, versionToLoad = versionToLoad, includeMinorCompactions = includeMinorCompactions) getLogSegmentForVersion( versionToLoad, newFiles, oldCheckpointProviderOpt = oldCheckpointProviderOpt, lastCheckpointInfo = lastCheckpointInfo )
Therefore, whenever a new commit appears, Delta Lake can see it and include new files in the joined dataset without any explicit action nor atomicity concern. If you need to combine a streaming and static datasets in Apache Spark Structured Streaming with some specific refresh needs, a table file format is the way to go!
Consulting

With nearly 17 years of experience, including 9 as data engineer, I offer expert consulting to design and optimize scalable data solutions.
As an O’Reilly author, Data+AI Summit speaker, and blogger, I bring cutting-edge insights to modernize infrastructure, build robust pipelines, and
drive data-driven decision-making. Let's transform your data challenges into opportunities—reach out to elevate your data engineering game today!
👉 contact@waitingforcode.com
🔗 past projects


