Last time I introduced Py4j which is the bridge between Apache Spark JVM codebase and Python client applications. Today it's a great moment to take a deeper look at their interaction in the context of data processing defined with the RDD and DataFrame APIs.
Data Engineering Design Patterns

Looking for a book that defines and solves most common data engineering problems? I wrote
one on that topic! You can read it online
on the O'Reilly platform,
or get a print copy on Amazon.
I also help solve your data engineering problems 👉 contact@waitingforcode.com 📩
DataFrame and RDD
When you define a DataFrame-based transformation, such as a filter method or aggregate, PySpark delegates its computation to the JVM DataFrame. How? Let's check the snippet from dataframe.py:
class DataFrame(PandasMapOpsMixin, PandasConversionMixin):
def __init__(self, jdf, sql_ctx):
self._jdf = jdf
self.sql_ctx = sql_ctx
# ...
def createOrReplaceTempView(self, name):
self._jdf.createOrReplaceTempView(name)
def exceptAll(self, other):
return DataFrame(self._jdf.exceptAll(other._jdf), self.sql_ctx)
def limit(self, num):
jdf = self._jdf.limit(num)
return DataFrame(jdf, self.sql_ctx)
def coalesce(self, numPartitions):
return DataFrame(self._jdf.coalesce(numPartitions), self.sql_ctx)
def filter(self, condition):
if isinstance(condition, str):
jdf = self._jdf.filter(condition)
elif isinstance(condition, Column):
jdf = self._jdf.filter(condition._jc)
else:
raise TypeError("condition should be string or Column")
return DataFrame(jdf, self.sql_ctx)
As you can see, PySpark's DataFrame has a private _jdf property which is an instance of Py4j's JavaObject referencing DataFrame instance on the JVM side. A JavaObject gives the possibility to call methods and access fields of the JVM counterpart. It means that whenever you invoke a Python's DataFrame method, you automatically invoke Scala's DataFrame. Therefore, calling a function doesn't involve exchanging the data whatsoever. The physical execution of the function happens on the JVM side.
RDD does the opposite. This API involves 2-way synchronization between the Python and JVM processes where the data gets stored on the JVM side but the computation happens in the Python interpreter. Below you can see a simplified example for the map(...) transformation where PythonRDD physically store the data got from the applied map function and 2 serialize/deserialize operations.
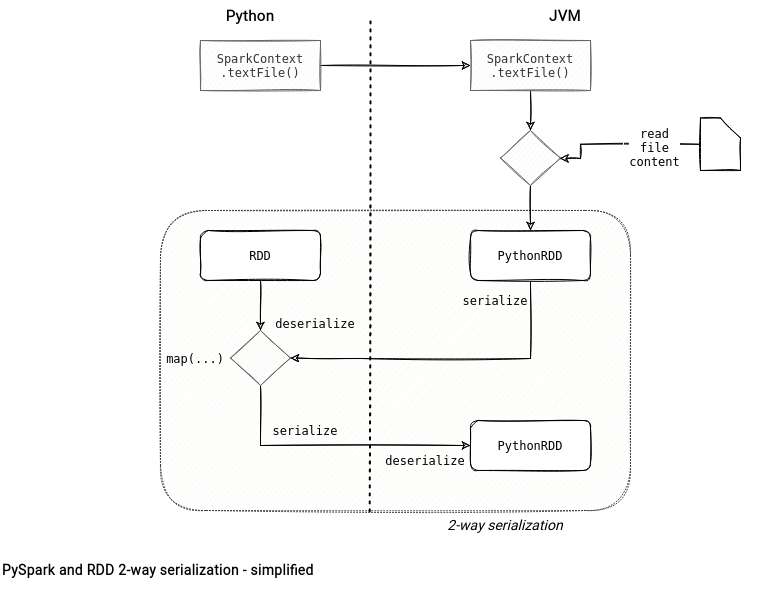
RDD in details
Let's take a deeper dive at the code used to draw the diagram in the previous section:
file_path = "/tmp/test_data.txt" text_file_rdd = spark.sparkContext.textFile(file_path, 2)
Python:The call gets delegated to the JVM's SparkContext:
class SparkContext(object): def textFile(self, name, minPartitions=None, use_unicode=True): minPartitions = minPartitions or min(self.defaultParallelism, 2) return RDD(self._jsc.textFile(name, minPartitions), self, UTF8Deserializer(use_unicode))JVM: The textFile() method loads the file content to an RDD.
-
def map_letter(letter: str) -> str: print(f'Mapping a {letter}') return f'{letter*2}x' mapped_text_lines = text_file_rdd.map(map_letter)Python: An RDD representing the textFile(...) result exists and it references its JVM counterpart. Internally, the map(...) call:
- Wraps the map_letter function with a wrapper failing on the stop iteration event:
class RDD(object): def map(self, f, preservesPartitioning=False): def func(_, iterator): return map(fail_on_stopiteration(f), iterator) return self.mapPartitionsWithIndex(func, preservesPartitioning) - Creates a new RDD of PipelineRDD type:
class RDD(object): def mapPartitionsWithIndex(self, f, preservesPartitioning=False): return PipelinedRDD(self, f, preservesPartitioning) -
Creates a new PythonRDD on the JVM side whenever any of the child RDD accesses the PipelineRDD's _jrdd property:
class PipelinedRDD(RDD): @property def _jrdd(self): # ... python_rdd = self.ctx._jvm.PythonRDD(self._prev_jrdd.rdd(), wrapped_func, self.preservesPartitioning, self.is_barrier) self._jrdd_val = python_rdd.asJavaRDD() if profiler: self._id = self._jrdd_val.id() self.ctx.profiler_collector.add_profiler(self._id, profiler) return self._jrdd_val
JVM: When the PipelineRDD computation triggers, it creates a new PythonRDD on the JVM side with the link to the parent RDD, the prepared function (wrapped with serializers).
The JVM takes the lines of the text file, serializes them to Pickle and sends to the Python process through the Py4j socket. - Wraps the map_letter function with a wrapper failing on the stop iteration event:
-
def map_letter(letter: str) -> str: print(f'Mapping a {letter}') return f'{letter*2}x' mapped_text_lines = text_file_rdd.map(map_letter)Python: The Python worker gets the serialized input, deserializes it, and passes it to the map_letter function. The interpreter serializes the outcome and sends it to the JVM through the socket.
JVM: The new process deserializes the incoming stream of transformed records and creates a new RDD.
mapped_text_lines.saveAsTextFile('/tmp/test_data_output')Python: Once again, PySpark delegates the physical execution to the JVM:
class RDD(object): def saveAsTextFile(self, path, compressionCodecClass=None): def func(split, iterator): for x in iterator: if not isinstance(x, (str, bytes)): x = str(x) if isinstance(x, str): x = x.encode("utf-8") yield x keyed = self.mapPartitionsWithIndex(func) keyed._bypass_serializer = True if compressionCodecClass: compressionCodec = self.ctx._jvm.java.lang.Class.forName(compressionCodecClass) keyed._jrdd.map(self.ctx._jvm.BytesToString()).saveAsTextFile(path, compressionCodec) else: keyed._jrdd.map(self.ctx._jvm.BytesToString()).saveAsTextFile(path)JVM: To see what happens the easiest way is to throw an exception by, for example, writing the same file twice. Below you'll find the stack trace for this scenario showing the reflection-based Py4j methods involved in the saveAsText code execution:
py4j.protocol.Py4JJavaError: An error occurred while calling o46.saveAsTextFile. : org.apache.hadoop.mapred.FileAlreadyExistsException: Output directory file:/tmp/test_data_output already exists # ... at org.apache.spark.rdd.RDD.saveAsTextFile(RDD.scala:1564) at org.apache.spark.api.java.JavaRDDLike.saveAsTextFile(JavaRDDLike.scala:551) at org.apache.spark.api.java.JavaRDDLike.saveAsTextFile$(JavaRDDLike.scala:550) at org.apache.spark.api.java.AbstractJavaRDDLike.saveAsTextFile(JavaRDDLike.scala:45) at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method) at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62) at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43) at java.lang.reflect.Method.invoke(Method.java:498) at py4j.reflection.MethodInvoker.invoke(MethodInvoker.java:244) at py4j.reflection.ReflectionEngine.invoke(ReflectionEngine.java:357) at py4j.Gateway.invoke(Gateway.java:282) at py4j.commands.AbstractCommand.invokeMethod(AbstractCommand.java:132) at py4j.commands.CallCommand.execute(CallCommand.java:79) at py4j.ClientServerConnection.waitForCommands(ClientServerConnection.java:182) at py4j.ClientServerConnection.run(ClientServerConnection.java:106) at java.lang.Thread.run(Thread.java:748)This last step doesn't involve any data movement towards the Python interpreter.
Why is map(...) not available in DataFrame?
PySpark DataFrame and RDD share several methods, like filter(...). It's one of the most common data transformations and from that we could naturally assume that another popular operation, the map(...), should also be present in both APIs. But this function is only available in the RDD.
One of the reasons might be the processing character. The filter(...) supports fully structured operation either on a Column or a string condition. It's not the case of the map(...) that can have any arbitrary processing logic. Hence, it must run on the Python side and get the data deserialized, which is the domain of the RDD.
Of course, the lack of the map function also comes from the dynamic vs static typing language and strongly typed character of Datasets. The filter method available in PySpark doesn't accept a custom function neither.
Serialization in the DataFrame
Although the DataFrame API is not supposed to serialize/deserialize the data from the JVM, it's not a rule. Any DataFrame method that brings the data to the driver needs to ask its Java counterpart to serialize it beforehand. Let's check some of these methods:
class DataFrame(PandasMapOpsMixin, PandasConversionMixin):
def collect(self):
with SCCallSiteSync(self._sc) as css:
sock_info = self._jdf.collectToPython()
return list(_load_from_socket(sock_info, BatchedSerializer(PickleSerializer())))
def toLocalIterator(self, prefetchPartitions=False):
with SCCallSiteSync(self._sc) as css:
sock_info = self._jdf.toPythonIterator(prefetchPartitions)
return _local_iterator_from_socket(sock_info, BatchedSerializer(PickleSerializer()))
def tail(self, num):
with SCCallSiteSync(self._sc):
sock_info = self._jdf.tailToPython(num)
return list(_load_from_socket(sock_info, BatchedSerializer(PickleSerializer())))
All of these 3 methods have a similar workflow consisting of loading the data from the socket and deserializing it to the Python objects. They all call a Scala Dataset API that registers the available serializers and converts them while returning the rows to the Python interpereter:
class Dataset[T] private[sql](
@DeveloperApi @Unstable @transient val queryExecution: QueryExecution,
@DeveloperApi @Unstable @transient val encoder: Encoder[T])
extends Serializable {
private[sql] def collectToPython(): Array[Any] = {
EvaluatePython.registerPicklers()
withAction("collectToPython", queryExecution) { plan =>
val toJava: (Any) => Any = EvaluatePython.toJava(_, schema)
val iter: Iterator[Array[Byte]] = new SerDeUtil.AutoBatchedPickler(
plan.executeCollect().iterator.map(toJava))
PythonRDD.serveIterator(iter, "serve-DataFrame")
}
}
The 2-way serialization mentioned many times in the article is often the main reason why it's recommended to prefer the DataFrame API over the RDD's. I hope that like me, now you understand better what happens and why it has such a big impact on the performance.
Consulting

With nearly 17 years of experience, including 9 as data engineer, I offer expert consulting to design and optimize scalable data solutions.
As an O’Reilly author, Data+AI Summit speaker, and blogger, I bring cutting-edge insights to modernize infrastructure, build robust pipelines, and
drive data-driven decision-making. Let's transform your data challenges into opportunities—reach out to elevate your data engineering game today!
👉 contact@waitingforcode.com
🔗 past projects


