Several weeks ago when I was checking new "apache-spark" tagged questions on StackOverflow I found one that caught my attention. The author was saying that randomSplit method doesn't divide the dataset equally and after merging back, the number of lines was different. Even though I wasn't able to answer at that moment, I decided to investigate this function and find possible reasons for that error.
Data Engineering Design Patterns

Looking for a book that defines and solves most common data engineering problems? I wrote
one on that topic! You can read it online
on the O'Reilly platform,
or get a print copy on Amazon.
I also help solve your data engineering problems 👉 contact@waitingforcode.com 📩
The post is composed of 2 parts. The first one describes the implementation of randomSplit function. In the second one I'm trying to show the logic behind this method.
Implementation details
Before I describe the internals of randomSplit method, I would like to start by its signature. The function randomly splits given dataset according to 2 parameters, an array of weights and a seed number. The weights are used to generate sample boundaries whereas seed ensures that the sampling on the same dataset executed several times will always return the same results:
"seed" should "always return the same samples" in {
val dataset = Seq(1, 2, 3, 4, 5).map(nr => nr).toDF("number")
val splitsForSeed11Run1 = dataset.randomSplit(Array(0.1, 0.1), seed = 11L)
val splitsForSeed11Run2 = dataset.randomSplit(Array(0.1, 0.1), seed = 11L)
val splitsForSeed12Run1 = dataset.randomSplit(Array(0.1, 0.1), seed = 12L)
def mapRowToInt(row: Row) = row.getAs[Int]("number")
val seed11Split0 = splitsForSeed11Run1(0).collect().map(mapRowToInt)
val seed11Split1 = splitsForSeed11Run1(1).collect().map(mapRowToInt)
splitsForSeed11Run2(0).collect().map(mapRowToInt) shouldEqual seed11Split0
splitsForSeed11Run2(1).collect().map(mapRowToInt) shouldEqual seed11Split1
splitsForSeed12Run1(0).collect().map(mapRowToInt) should not equal seed11Split0
splitsForSeed12Run1(1).collect().map(mapRowToInt) should not equal seed11Split1
}
randomSample starts by generating sort expression directly from the columns returned by the logical plan. The goal is to make the ordering deterministic and therefore, avoid the risk of overlapping splits. If the columns cannot be sorted, the ordering is replaced by Dataset materialization through a cache. I will focus more on this determinism in the next section because it's the key for the randomSplit logic.
After that, if the sum of weights is not equal to 1, Apache Spark computes normalized cumulative weights that will be used to generate splitted Datasets:
val sum = weights.sum
val normalizedCumWeights = weights.map(_ / sum).scanLeft(0.0d)(_ + _)
normalizedCumWeights.sliding(2).map { x =>
new Dataset[T](
sparkSession, Sample(x(0), x(1), withReplacement = false, seed, plan), encoder)
}.toArray
Datasets are constructed directly from a logical plan starting with Sample operator which at the moment of execution is handled by org.apache.spark.sql.execution.SampleExec physical plan. Later, for this operator (not randomSplut) the data is generated with one of available samplers, BernoulliCellSampler or PoissonSampler.
Two samplers? Indeed, but they're used for specific cases. PoissonSampler is used when the sampling is configured with a withReplacement parameter set to true. For randomSplit, this parameter is always false, but it's worth some explanation. Replacement in the context of sampling comes from statistics. Sampling with replacement is the sampling when a unit can occur one or more times in the sample. Sampling without replacement is the opposite. So for our randomSplit use case where the withReplacement is false, BernoulliCellSampler will be used.
Execution details
Using randomSplit, as you saw it already, is straightforward. You need to define the weights and the seed for reproducibility. And as you already know, weights don't need to be equal to 1:
"randomSplit" should "return 2 splits with normalized weights" in {
val dataset = (1 to 10).map(nr => nr).toDF("number")
val splits = dataset.randomSplit(Array(0.1, 0.1), seed = 11L)
val split1 = splits(0)
split1.explain(true)
val split1Data = split1.collect().map(mapRowToInt)
split1Data should have size 6
val split2 = splits(1)
split2.explain(true)
val split2Data = split2.collect().map(mapRowToInt)
split2Data should have size 4
}
As you can see, I defined weights whose sum is not equal to 1. If you read the previous section carefully, you've certainly noticed the weight normalization step if the sum is not 1. It happens here where the normalized weights are (0, 0.5, 1) rather than (0.1, 0.1, 1). You can also notice that by analyzing execution plans:
# split1 == Physical Plan == *(1) Sample 0.0, 0.5, false, 11 +- *(1) Sort [number#3 ASC NULLS FIRST], false, 0 +- LocalTableScan [number#3] # split2 == Physical Plan == *(1) Sample 0.5, 1.0, false, 11 +- *(1) Sort [number#3 ASC NULLS FIRST], false, 0 +- LocalTableScan [number#3]
I show you the execution plan for one purpose - explaining the internal execution. As you can see, in our test case everything begins with a sort. If so, why you don't see a shuffle exchange in the plan? After all, sorting a dataset should involve such an action. The point is that the Sort used here is not global sort and you can see that with the false parameter. The ordering is then local to each partition and you can also notice that by analyzing the generated code:
/* 045 */ protected void processNext() throws java.io.IOException {
/* 046 */ if (sort_needToSort_0) {
/* 047 */ long sort_spillSizeBefore_0 = sort_metrics_0.memoryBytesSpilled();
/* 048 */ sort_addToSorter_0();
/* 049 */ sort_sortedIter_0 = sort_sorter_0.sort();
/* 050 */ ((org.apache.spark.sql.execution.metric.SQLMetric) references[3] /* sortTime */).add(sort_sorte
r_0.getSortTimeNanos() / 1000000);
/* 051 */ ((org.apache.spark.sql.execution.metric.SQLMetric) references[1] /* peakMemory */).add(sort_sor
ter_0.getPeakMemoryUsage());
/* 052 */ ((org.apache.spark.sql.execution.metric.SQLMetric) references[2] /* spillSize */).add(sort_metr
ics_0.memoryBytesSpilled() - sort_spillSizeBefore_0);
/* 053 */ sort_metrics_0.incPeakExecutionMemory(sort_sorter_0.getPeakMemoryUsage());
/* 054 */ sort_needToSort_0 = false;
/* 055 */ }
/* 056 */
/* 057 */ while (sort_sortedIter_0.hasNext()) {
/* 058 */ UnsafeRow sort_outputRow_0 = (UnsafeRow)sort_sortedIter_0.next();
/* 059 */
/* 060 */ if (sample_mutableStateArray_0[0].sample() != 0) {
/* 061 */ ((org.apache.spark.sql.execution.metric.SQLMetric) references[4] /* numOutputRows */).add(1);
/* 062 */
/* 063 */ int sort_value_0 = sort_outputRow_0.getInt(0);
/* 064 */ boolean project_isNull_0 = false;
/* 065 */ UTF8String project_value_0 = null;
/* 066 */ if (!false) {
/* 067 */ project_value_0 = UTF8String.fromString(String.valueOf(sort_value_0));
/* 068 */ }
/* 069 */ sample_mutableStateArray_1[1].reset();
/* 070 */
/* 071 */ sample_mutableStateArray_1[1].zeroOutNullBytes();
/* 072 */
/* 073 */ sample_mutableStateArray_1[1].write(0, project_value_0);
/* 074 */ append((sample_mutableStateArray_1[1].getRow()));
/* 075 */
/* 076 */ }
Fine, but my previous example contained already ordered items. What happens if they're unordered, like here?
val dataset = Seq(1, 3, 4, 5, 2, 7, 10, 9, 8, 11, 20, 18, 17, 16).map(nr => nr).toDF("number")
val splits = dataset.randomSplit(Array(0.1, 0.1), seed = 11L)
val split1 = splits(0)
split1.show(true)
val split2 = splits(1)
split2.show(true)
This time, I will answer that with an image:
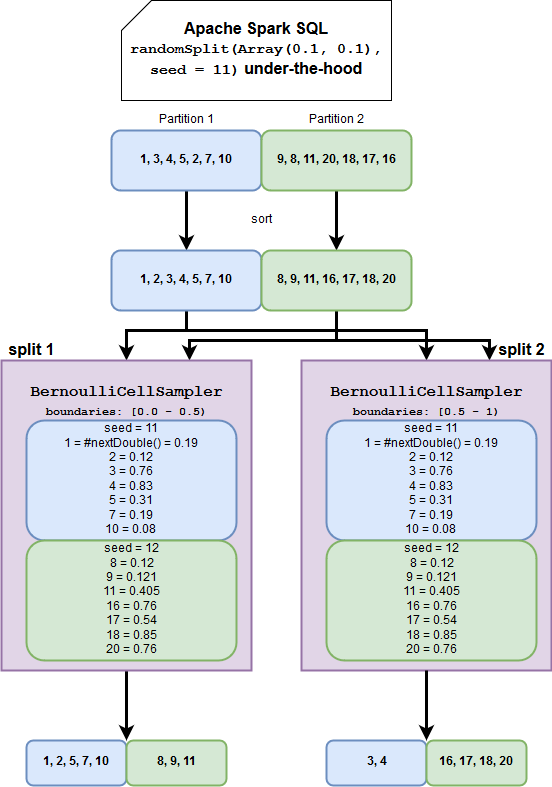
Two things to note here. The first one is the importance of the seed. It remains the same for 2 splits and that's why the rows aren't duplicated. The second one is the physical execution. The input dataset partitions are processed twice, once for each split. It explains why Apache Spark doesn't need to shuffle data in order to guarantee sampling consistency. It also shows the importance of ordering. Alongside the seed and used random generator (XORShiftRandom), ordering guarantees determinism. If you analyze the image, you will see that the rows are always processed in the same order and that the random generator always generates the same number for the given line. If the data weren't ordered, we would end up with duplicates or missing rows.
Seed is also important for XORShiftRandom called by BernouliCellSampler because it uses the initial one and creates a new one before each sample in deterministic manner:
private var seed = XORShiftRandom.hashSeed(init)
// we need to just override next - this will be called by nextInt, nextDouble,
// nextGaussian, nextLong, etc.
override protected def next(bits: Int): Int = {
var nextSeed = seed ^ (seed << 21)
nextSeed ^= (nextSeed >>> 35)
nextSeed ^= (nextSeed << 4)
seed = nextSeed
(nextSeed & ((1L << bits) -1)).asInstanceOf[Int]
}
// It's called from Random.java
// public double nextDouble() {
// return (((long)(next(26)) << 27) + next(27)) * DOUBLE_UNIT;
// }
At first contact, randomSplit scared me a little. It talked about sampling with replacement, without replacement, determinism requirement and some other concepts that I was not familiar with. Fortunately, some tests and debugging breakpoints later, I was able to understand it in a big picture. I hope that you too and in case of any issue with randomSplit, this post will help you to figure out the reason faster. BTW, if you have any code making randomSplit behave differently than it should be, do not hesitate to share it with the community. There were some cases in the past (SPARK-12662) and maybe there are still some corner cases uncovered?
Consulting

With nearly 17 years of experience, including 9 as data engineer, I offer expert consulting to design and optimize scalable data solutions.
As an O’Reilly author, Data+AI Summit speaker, and blogger, I bring cutting-edge insights to modernize infrastructure, build robust pipelines, and
drive data-driven decision-making. Let's transform your data challenges into opportunities—reach out to elevate your data engineering game today!
👉 contact@waitingforcode.com
🔗 past projects


