https://github.com/bartosz25/data-ai-summit-2024
With this blog I'm starting a follow-up series for my Data+AI Summit 2024 talk. I missed this family of blog posts a lot as the previous DAIS with me as speaker was 4 years ago! As previously, this time too I'll be writing several blog posts that should help you remember the talk and also cover some of the topics left aside because of the time constraints.
Data Engineering Design Patterns

Looking for a book that defines and solves most common data engineering problems? I wrote
one on that topic! You can read it online
on the O'Reilly platform,
or get a print copy on Amazon.
I also help solve your data engineering problems 👉 contact@waitingforcode.com 📩
The first blog post of the series introduces a method that you can use to structure your Apache Spark Structured Streaming project in a testable and readable manner. But before you see it in action, I owe you some theoretical introduction first.
The Dataflow Model: A Practical Approach to Balancing Correctness, Latency, and Cost in Massive-Scale, Unbounded, Out-of-Order Data Processing
The authors of the paper are Tyler Akidau, Robert Bradshaw, Craig Chambers, Slava Chernyak, Rafael J. Fernandez-Moctezuma, Reuven Lax, Sam McVeety, Daniel Mills, Frances Perry, Eric Schmidt, and Sam Whittle.
Dataflow model
The structure idea comes from the Dataflow model which is a concept formalized in 2015 by Google engineers in a paper called "The Dataflow Model: A Practical Approach to Balancing Correctness, Latency, and Cost in Massive-Scale, Unbounded, Out-of-Order Data Processing". If you don't want to read the paper, you can also take a look at Streaming: The world beyond batch, part 1 and part 2 series written by Tyler Akidau, who is one of the authors of the original paper.
That's for the context, but what is this Dataflow model? Although it's more that the things I'll show you here, let's stay short and clear and focus on the essentials that might help structure an Apache Spark Structured Streaming code. One of Dataflow important parts is the description of a streaming pipeline by 4 dimensions: What, Where, When, and How. Each dimension is responsible for a dedicated technical component of a streaming pipeline. To be more precise, let's see the table below that by the way, also explains Apache Spark Structured Streaming integration:
| Dataflow component | Explanation as per "The Dataflow Model: A..." | Structured Streaming application |
|---|---|---|
| What | results are being computed. | The business logic you define supported by the available Apache Spark operators. |
| Where | in event time the results are being computed. | Watermark, if the stateful results are accumulated. |
| When | in processing time the results are materialized. | Micro-batch and processing time trigger. |
| How | earlier results relate to later refinements aka how new records interact with the already accumulated ones, especially for the stateful processing. | Output modes, particularly the Update which combines new records with the old ones. |
You can take a look at full capabilities in the Dataflow (now Beam) model matrix. I might have simplified some of them for Apache Spark Structured Streaming, but despite that, it's enough to go further and start working on our project structure.
Demo project introduction
To better explain the code organization workflow, we'll use an arbitrary stateful processing job that builds navigation sessions from raw input visits. I like this example because it's complex and simple at the same time. It's complex due to the extended business logic but it remains simple as it doesn't involve any fancy operations such as stream-to-stream joins, or multiple watermarks.
Our sample job will read raw visits from an Apache Kafka topic, extract important fields, and aggregated them as pending sessions. Once the session expires, the mapping function will generate the final output that we're going to join with devices enrichment dataset stores as a Delta Lake table. The logic overall looks like in the snippet below:
input_data_stream = ...
devices_table = ...
grouped_visits = (input_data_stream
.withWatermark('event_time', '1 minute').groupBy(F.col('visit_id')))
sessions = grouped_visits.applyInPandasWithState(
# ...
)
enriched_sessions = enrich_sessions_with_devices(sessions, devices_table)
visits_to_write = enriched_sessions.withColumn('value', F.to_json(F.struct('*'))).select('value')
visits_to_write.writeStream
.outputMode("append")
.trigger(processingTime='15 seconds')
.format("kafka").
# ...
Let's see now how to apply the Dataflow model's dimensions to our job.
Project organization and the Dataflow model
After applying the Dataflow model to our job, we should end up with the annotations like the ones from the next picture:
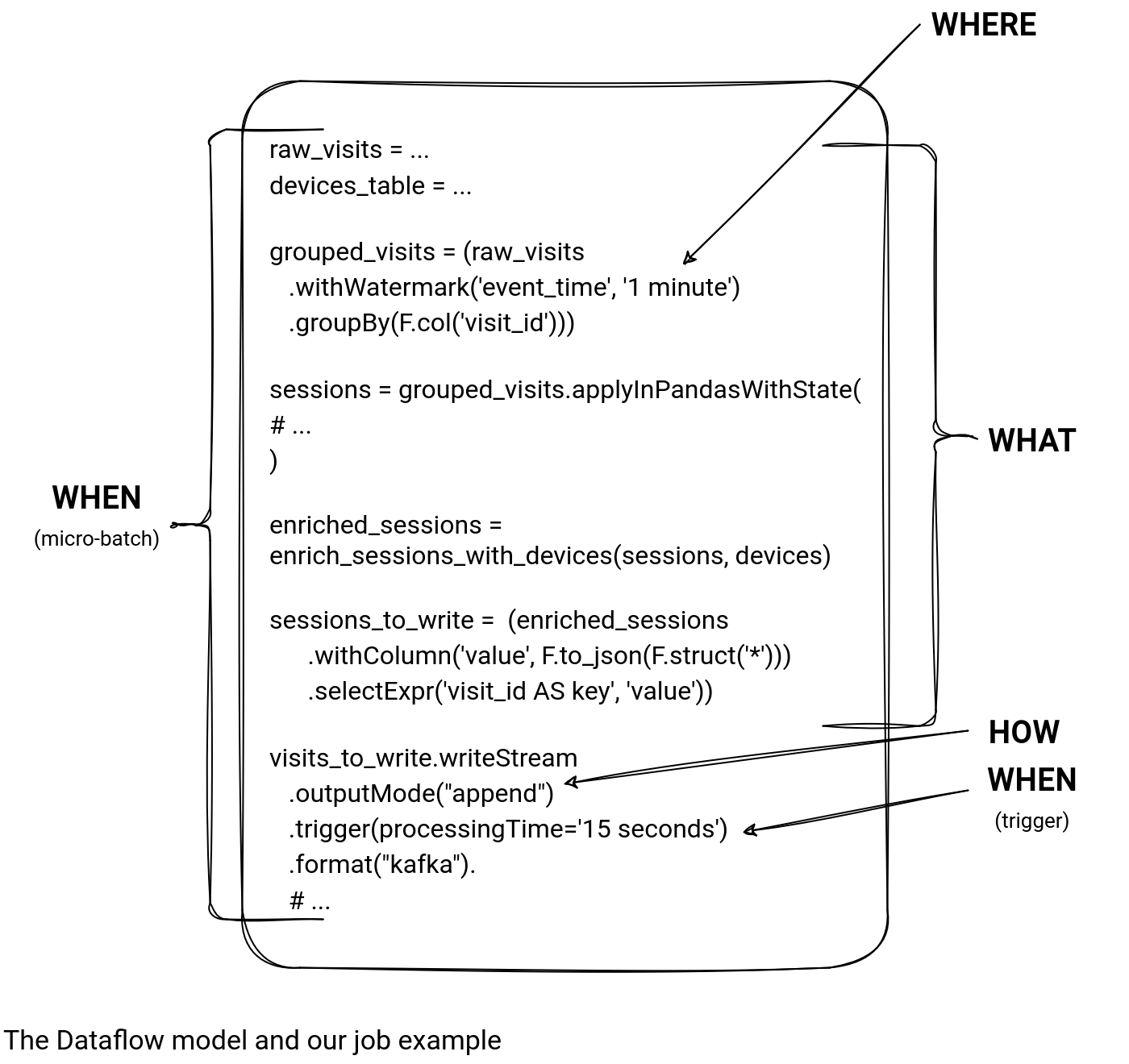
Let's deep dive into each of the dimensions:
- How. It's the easiest one as the single function dealing with the accumulated records is the outputMode. As you can see, our job handles late data by emitting only the final result after the watermark, i.e. it ignores any record being late compared to the given watermark.
- What. It's the definition of the processing logic, which in our case has several steps, like: the input record formatting with a dedicated schema, the stateful mapping preceded by the grouping, the join between static and dynamic DataFrames, and finally, the output generation.
- When. It points down to the micro-batch and trigger, both aspects impacting the results generation from the processing time standpoint.
- Where. It scopes the calculation in terms of event time, which in our case is controlled by the watermark.
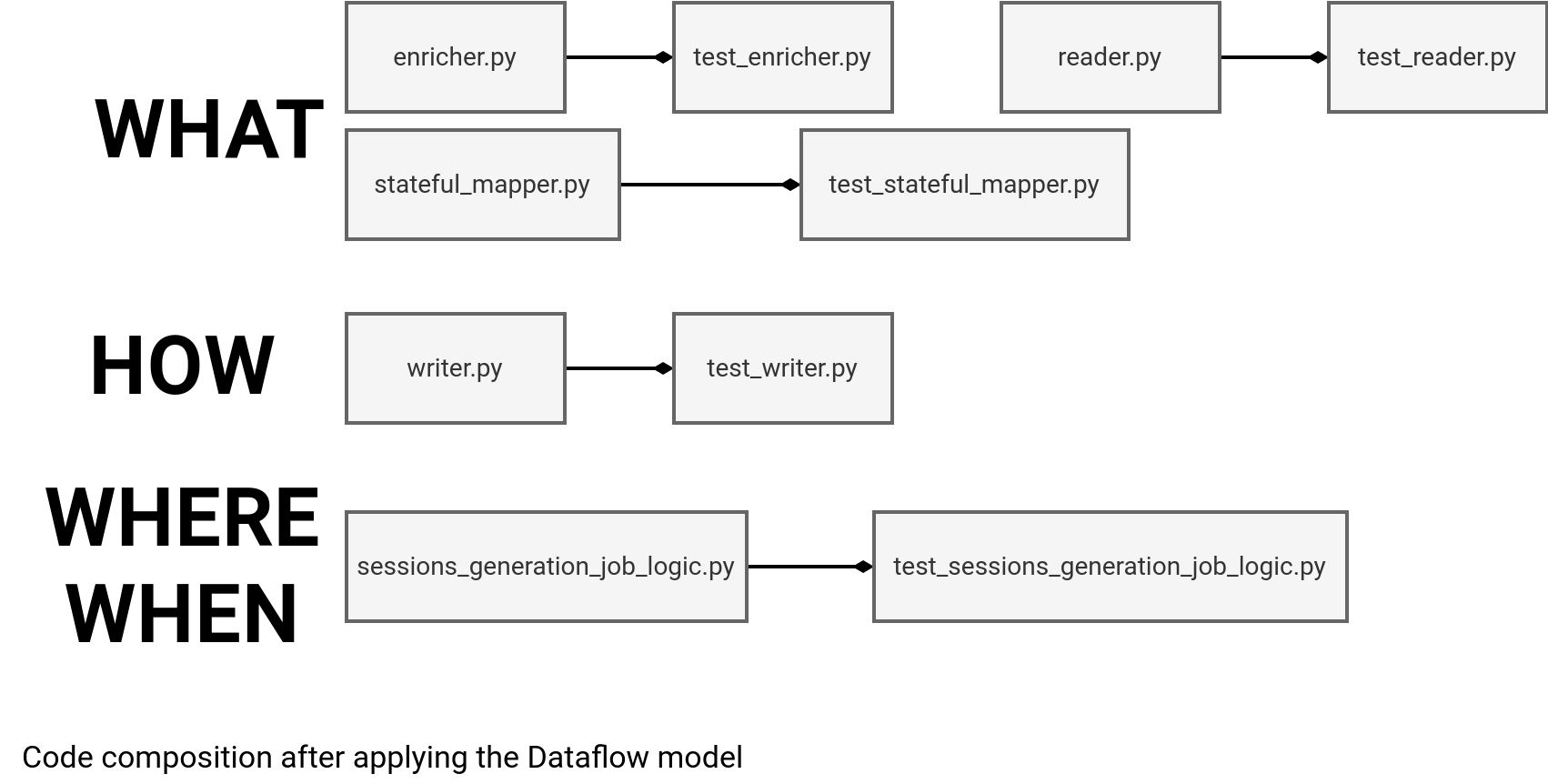
After this definition step, we can now divide the monolithic job from the first snippet into modules according to the Dataflow categories:
Implementation details
As the picture is great but the code is even better, let me dive into the implementation details and show you the operations present in each file.
The first one is the reader.py that takes a DataFrame, typically the Kafka data source's one, and extracts the columns. As you can see in the code just below, it contains our business logic since the job expects to extract only several columns from the whole row via the visit_schema variable:
def select_raw_visits(visits_source: DataFrame) -> DataFrame:
visit_schema = '''visit_id STRING, event_time TIMESTAMP, user_id STRING, page STRING,...'''
return (visits_source.select(
F.from_json(F.col('value').cast('string'), visit_schema).alias('value'))
.selectExpr('value.*'))
Now comes probably the most complex part, the sessionization logic in the stateful_mapper.py file. To start simple, you can validate the mapping function working in isolation, i.e. without iteration. That way you will validate the static behavior where you fully control the input:
def map_visits_to_session(visit_id_tuple: Any,
input_rows: Iterable[pandas.DataFrame],
current_state: GroupState) -> Iterable[pandas.DataFrame]:
session_expiration_time_10min_as_ms = 10 * 60 * 1000
visit_id = visit_id_tuple[0]
visit_to_return = None
if current_state.hasTimedOut:
# ...
visit_to_return = get_session_to_return(visit_id, visits, user_id)
else:
# ...
if visit_to_return:
yield pandas.DataFrame(visit_to_return)
After that, the next step is to validate the enrichment with the static Delta Lake table present in the enricher.py file. Although the join operation is relatively simple, you can already see that the code also acts as a specification for both the join condition and retained values:
def enrich_sessions_with_devices(sessions: DataFrame, devices: DataFrame) -> DataFrame:
enriched_visits = ((sessions.join(
devices,
[sessions.device_type == devices.type,
sessions.device_version == devices.version],
'left_outer')).drop('type', 'version', 'device_full_name')
.withColumnRenamed('full_name', 'device_full_name'))
return enriched_visits
I'm omitting here the tests on the sessions_to_write DataFrame, as it's a relatively straightforward operation. That said, adding a test here is not a crime, but skipping it isn't one either. And before we go to the last WHEN/WHERE part, we can also add a test to validate the output mode, hence the HOW, and ensure any change at this level will trigger the error:
def set_up_visits_writer(visits_to_write: DataFrame, trigger: Dict[str, str]) -> DataStreamWriter:
return (visits_to_write.writeStream
.outputMode("append")
.trigger(**trigger))
Tests granularity
As for the sessions_to_write, it's valid to skip the tests for the set_up_visits_writer function. In our example, the function is part of the sessions_generation_job_logic.py and will be tested at the moment of validating the whole sessionization logic. Once again, it boils down to you and your team practices. If you like validating the smallest units of work before asserting on the bigger ones, adding a simple test can be good. Otherwise, it won't be a crime if you consider it as an extra code to maintain that either way will be covered in a different place.
The remaining parts are the WHEN and WHERE. You can use them to validate the final, IO-agnostic, data processing logic present in the sessions_generation_job_logic.py:
def generate_sessions(visits_source: DataFrame, devices: DataFrame, trigger: Dict[str, str],
checkpoint_location: str) -> DataStreamWriter:
raw_visits = select_raw_visits(visits_source)
grouped_visits = (raw_visits
.withWatermark('event_time', '5 minutes')
.groupBy(F.col('visit_id')))
sessions = grouped_visits.applyInPandasWithState(
func=map_visits_to_session,
outputStructType=get_session_output_schema(),
stateStructType=get_session_state_schema(),
outputMode="append",
timeoutConf="EventTimeTimeout"
)
enriched_sessions = enrich_sessions_with_devices(sessions, devices)
sessions_to_write = (enriched_sessions
.withColumn('value', F.to_json(F.struct('*')))
.selectExpr('visit_id AS key', 'value'))
return set_up_sessions_writer(sessions_to_write, trigger).option('checkpointLocation', checkpoint_location)
Let's stop for a moment here. As you can see, the code doesn't return a DataFrame but the DataStreamWriter. Thanks to that, you can validate it against the consistent HOW condition defined in the output mode, and also have a possibility to materialize the output to any data store supported by Apache Spark. For our tests, we're going to use a custom in-memory store based on the foreachBatch(...) sink but that's the story for the next blog post!
Consulting

With nearly 17 years of experience, including 9 as data engineer, I offer expert consulting to design and optimize scalable data solutions.
As an O’Reilly author, Data+AI Summit speaker, and blogger, I bring cutting-edge insights to modernize infrastructure, build robust pipelines, and
drive data-driven decision-making. Let's transform your data challenges into opportunities—reach out to elevate your data engineering game today!
👉 contact@waitingforcode.com
🔗 past projects


