It's time to start the 4th part of the Table file formats series. This time the topic will be Change Data Capture, so how to stream all changes made on the table. As for the 3rd part, I'm going to start with Delta Lake.
Data Engineering Design Patterns

Looking for a book that defines and solves most common data engineering problems? I wrote
one on that topic! You can read it online
on the O'Reilly platform,
or get a print copy on Amazon.
I also help solve your data engineering problems 👉 contact@waitingforcode.com 📩
Change Data Feed 101
Change Data Capture in Delta Lake is also known as Change Data Feed. To enable it, you can either CREATE or ALTER TABLE with the delta.enableChangeDataFeed = true property. Besides, you can also apply this configuration globally, to all tables created in the given SparkSession. To do that, you should set the spark.databricks.delta.properties.defaults.enableChangeDataFeed to true at the session level.
Enabling the CDC (CDF in Delta Lake nomenclature, I'm going to keep the CDC for the sake of simplicity) implies different things that you will discover more in detail later in the blog post. But in a nutshell:
- Possibility to read data or the changes. The difference? There is an extra column and additional rows for the the changes feed lecture that can be enabled with the .option("readChangeFeed", "true"):
# readChangeFeed = true +---+-----+--------+----------------+---------------+--------------------+ | id|login|isActive| _change_type|_commit_version| _commit_timestamp| +---+-----+--------+----------------+---------------+--------------------+ | 4|user4| true| update_preimage| 1|2022-10-22 15:14:...| | 4|user4| false|update_postimage| 1|2022-10-22 15:14:...| | 6|user6| true| insert| 1|2022-10-22 15:14:...| | 1|user1| true| insert| 0|2022-10-22 15:13:...| | 2|user2| false| insert| 0|2022-10-22 15:13:...| | 3|user3| true| insert| 0|2022-10-22 15:13:...| | 4|user4| true| insert| 0|2022-10-22 15:13:...| | 5| | true| insert| 0|2022-10-22 15:13:...| +---+-----+--------+----------------+---------------+--------------------+ # readChangeFeed = false +---+-----+--------+ |id |login|isActive| +---+-----+--------+ |1 |user1|true | |2 |user2|false | |3 |user3|true | |4 |user4|false | |5 | |true | |6 |user6|true | +---+-----+--------+
- As a result of the above outcome, data writers have more work.
- This extra work brings a change on the data storage layout and commit log format too.
Even though it can be enough to start working with the CDC in Delta Lake, let's see what happens exactly when you write and read tables with the Change Feed enabled.
Writing 1 - operations semantics
Enabling the CDC involves heavier writing for the DELETE, MERGE, and UPDATE operations:
- DELETE and MERGE - DELETE. The output contains all rows, even the ones matching the delete condition. Otherwise, it wouldn't be possible to stream the delete changes.
- MERGE - UPDATE and UPDATE. The output has 2 rows. The first row stores the data before the operation and it's marked with the update_preimage change type. The second stores the data after the update and you can find it with the update_postimage type.
Having these extra rows to output involves more data to generate, hence a bigger memory and compute footprints in the application. Let me show you that in the code:
def withUpdatedColumns(
target: LogicalPlan, updateExpressions: Seq[Expression], condition: Expression,
dfWithEvaluatedCondition: DataFrame, shouldOutputCdc: Boolean): DataFrame = {
// ...
val namedUpdateCols = updateExpressions.zip(target.output).map {
case (expr, targetCol) => new Column(expr).as(targetCol.name)
}
val preimageCols = target.output.map(new Column(_)) :+
lit(CDC_TYPE_UPDATE_PREIMAGE).as(CDC_TYPE_COLUMN_NAME)
val postimageCols = namedUpdateCols :+
lit(CDC_TYPE_UPDATE_POSTIMAGE).as(CDC_TYPE_COLUMN_NAME)
val updatedDataCols = namedUpdateCols :+
typedLit[String](CDC_TYPE_NOT_CDC).as(CDC_TYPE_COLUMN_NAME)
val noopRewriteCols = target.output.map(new Column(_)) :+
typedLit[String](CDC_TYPE_NOT_CDC).as(CDC_TYPE_COLUMN_NAME)
val packedUpdates = array(
struct(preimageCols: _*),
struct(postimageCols: _*),
struct(updatedDataCols: _*)
).expr
val packedData = if (condition == Literal.TrueLiteral) {
packedUpdates
} else {
If(
UnresolvedAttribute(CONDITION_COLUMN_NAME),
packedUpdates, // if it should be updated, then use `packagedUpdates`
array(struct(noopRewriteCols: _*)).expr) // else, this is a noop rewrite
}
// Explode the packed array, and project back out the final data columns.
val finalColNames = target.output.map(_.name) :+ CDC_TYPE_COLUMN_NAME
dfWithEvaluatedCondition
.select(explode(new Column(packedData)).as("packedData"))
.select(finalColNames.map { n => col(s"packedData.`$n`").as(s"$n") }: _*)
The logic creates 4 column sets. Three of them (preimageCols, postimageCols, updatedDataCols) are used as the CDC output if the original row matches the update predicate. The last created row (nooprewriteCols) is returned when the predicate doesn't apply. As a consequence, each update for a CDC table generates 3 or 1 row.
The logic for a DELETE operation always returns all the rows with the really deleted rows annotated with a "delete" type in the _change_type column. So the overhead will be here the number of removed rows.
Finally, for the MERGE:
- the DELETE generates 2 rows for CDC or 1 row otherwise:
val mainDataOutput = targetOutputCols :+ TrueLiteral :+ incrDeletedCountExpr :+
Literal(CDC_TYPE_NOT_CDC)
if (cdcEnabled) {
// For delete we do a no-op copy with ROW_DELETED_COL = false, INCR_ROW_COUNT_COL as a
// no-op (because the metric will be incremented in `mainDataOutput`) and
// CDC_TYPE_COLUMN_NAME = CDC_TYPE_DELETE
val deleteCdcOutput = targetOutputCols :+ FalseLiteral :+ TrueLiteral :+
Literal(CDC_TYPE_DELETE)
Seq(mainDataOutput, deleteCdcOutput)
} else {
Seq(mainDataOutput)
}
- the UPDATE generates 3 rows for CDC or 1 otherwise
val mainDataOutput = u.resolvedActions.map(_.expr) :+ FalseLiteral :+
incrUpdatedCountExpr :+ Literal(CDC_TYPE_NOT_CDC)
if (cdcEnabled) {
// For update preimage, we have do a no-op copy with ROW_DELETED_COL = false and
// CDC_TYPE_COLUMN_NAME = CDC_TYPE_UPDATE_PREIMAGE and INCR_ROW_COUNT_COL as a no-op
// (because the metric will be incremented in `mainDataOutput`)
val preImageOutput = targetOutputCols :+ FalseLiteral :+ TrueLiteral :+
Literal(CDC_TYPE_UPDATE_PREIMAGE)
// For update postimage, we have the same expressions as for mainDataOutput but with
// INCR_ROW_COUNT_COL as a no-op (because the metric will be incremented in
// `mainDataOutput`), and CDC_TYPE_COLUMN_NAME = CDC_TYPE_UPDATE_POSTIMAGE
val postImageOutput = mainDataOutput.dropRight(2) :+ TrueLiteral :+
Literal(CDC_TYPE_UPDATE_POSTIMAGE)
Seq(mainDataOutput, preImageOutput, postImageOutput)
} else {
Seq(mainDataOutput)
}
- the INSERT case generates one extra row to the initially added record:
if (cdcEnabled) {
// For insert we have the same expressions as for mainDataOutput, but with
// INCR_ROW_COUNT_COL as a no-op (because the metric will be incremented in
// `mainDataOutput`), and CDC_TYPE_COLUMN_NAME = CDC_TYPE_INSERT
val insertCdcOutput = mainDataOutput.dropRight(2) :+ TrueLiteral :+ Literal(CDC_TYPE_INSERT)
Seq(mainDataOutput, insertCdcOutput)
} else {
Seq(mainDataOutput)
}
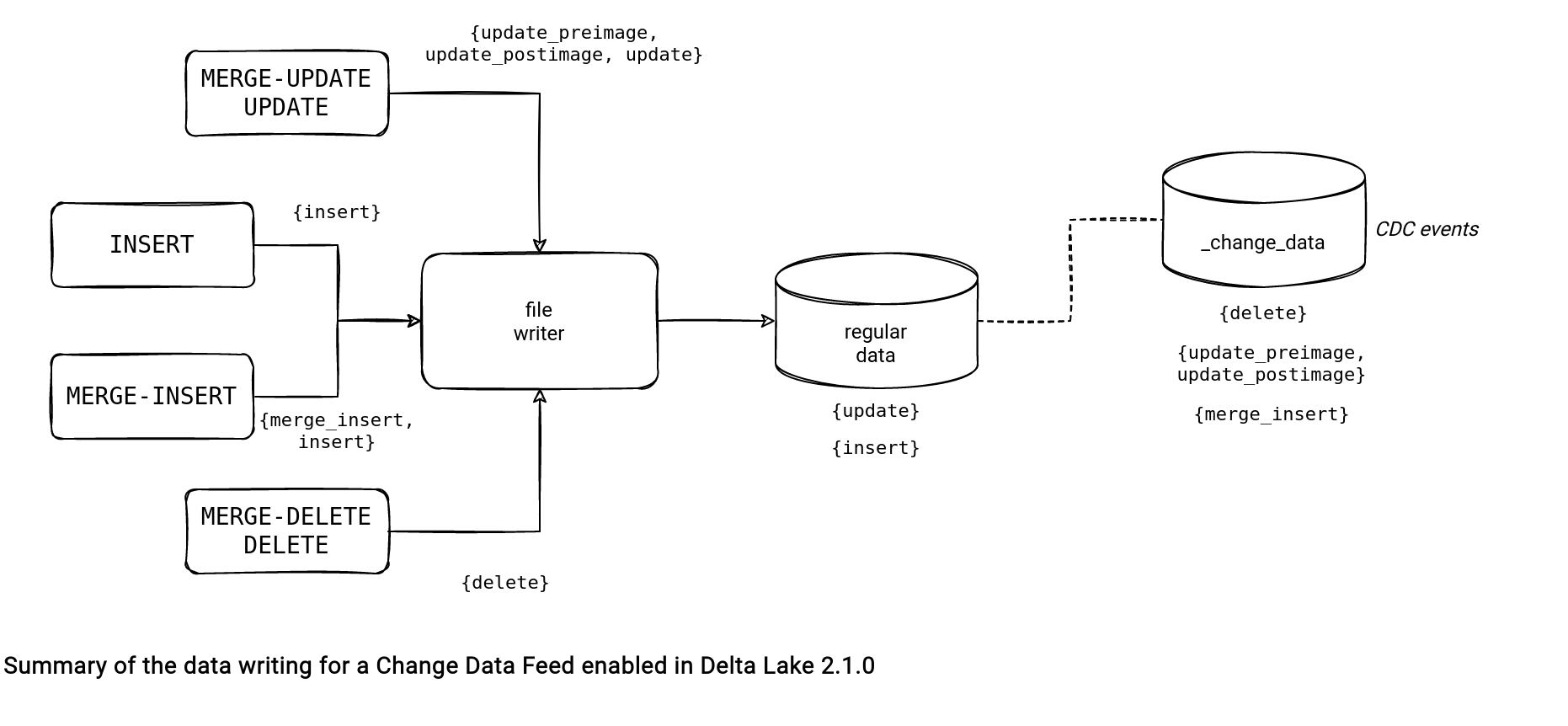
You may have noticed the absence of the INSERT operation in the list. It's normal since this event doesn't require any special treatment in terms of CDC. The file generated by the writer contains only the added records ("insert" CDC type) and at reading, Delta Lake can simply take it and return to the consumer. It doesn't happen for the MERGE-INSERT, though, because this operation is executed alongside in-place changes and it's very probable, it won't generate a file with new data only. You should be able to understand why after the reading part of this blog post.
Writing 2 - storage and commit log
Once the output DataFrame is generated, Delta Lake takes care of writing them to the files. The writing process doesn't put all rows in the same location. Instead, it splits the CDC events from the regular rows and writes them to the files located in the _change_data directory. The part responsible for this split is the performCDCPartition(inputData: Dataset[_]):
trait TransactionalWrite extends DeltaLogging { self: OptimisticTransactionImpl =>
// ...
protected def performCDCPartition(inputData: Dataset[_]): (DataFrame, StructType) = {
if (CDCReader.isCDCEnabledOnTable(metadata) &&
inputData.schema.fieldNames.contains(CDCReader.CDC_TYPE_COLUMN_NAME)) {
val augmentedData = inputData.withColumn(
CDCReader.CDC_PARTITION_COL, col(CDCReader.CDC_TYPE_COLUMN_NAME).isNotNull)
val partitionSchema = StructType(
StructField(CDCReader.CDC_PARTITION_COL, StringType) +: metadata.physicalPartitionSchema)
(augmentedData, partitionSchema)
} else {
(inputData.toDF(), metadata.physicalPartitionSchema)
}
}
The code snippet adds an extra column for the CDC-related events (_change_type column defined). The DelayedCommitProtocol uses that column later to create either regular or CDC task output file:
class DelayedCommitProtocol(jobId: String, path: String, randomPrefixLength: Option[Int])
extends FileCommitProtocol with Serializable with Logging {
// ...
protected def getFileName(taskContext: TaskAttemptContext, ext: String, partitionValues: Map[String, String]): String = {
// The file name looks like part-r-00000-2dd664f9-d2c4-4ffe-878f-c6c70c1fb0cb_00003.gz.parquet
// Note that %05d does not truncate the split number, so if we have more than 100000 tasks,
// the file name is fine and won't overflow.
val split = taskContext.getTaskAttemptID.getTaskID.getId
val uuid = UUID.randomUUID.toString
// CDC files (CDC_PARTITION_COL = true) are named with "cdc-..." instead of "part-...".
if (partitionValues.get(CDC_PARTITION_COL).contains("true")) {
f"cdc-$split%05d-$uuid$ext"
} else {
f"part-$split%05d-$uuid$ext"
}
}
The final step moves these cdc- files to the _change_data directory introduced previously:
override def newTaskTempFile(
// ...
val filename = getFileName(taskContext, ext, partitionValues)
// ...
if (subDir == cdcPartitionFalse) {
new Path(filename)
} else if (subDir.startsWith(cdcPartitionTrue)) {
val cleanedSubDir = cdcPartitionTrueRegex.replaceFirstIn(subDir, CDC_LOCATION)
new Path(cleanedSubDir, filename)
///...
Indeed, all this logic must be reflected in the commit log that contains an extra "cdc" action, as in the example below:
{"remove":{"path":"part-00000-67f642c5-f62a-4e7e-ad19-42eaefd7716e-c000.snappy.parquet","deletionTimestamp":1666496845341,"dataChange":true,"extendedFileMetadata":true,"partitionValues":{},"size":924}}
{"add":{"path":"part-00000-4014c3a8-45b7-41f9-b236-efc2680d3975.c000.snappy.parquet","partitionValues":{},"size":1145,"modificationTime":1666496845312,"dataChange":true,"stats":"{\"numRecords\":4,\"minValues\":{\"id\":1,\"login\":\"user1\"},\"maxValues\":{\"id\":4,\"login\":\"user4\"},\"nullCount\":{\"id\":0,\"login\":0,\"isActive\":0,\"_change_type\":4}}"}}
{"cdc":{"path":"_change_data/cdc-00000-1d388b15-9cb1-48f2-8ce9-ea14b528cb9a.c000.snappy.parquet","partitionValues":{},"size":1148,"dataChange":false}}
{"commitInfo":{"timestamp":1666496845363,"operation":"DELETE","operationParameters":{"predicate":"[\"(spark_catalog.default.rateDataInPlace.login = '')\"]"},"readVersion":0,"isolationLevel":"Serializable","isBlindAppend":false,"operationMetrics":{"numRemovedFiles":"1","numCopiedRows":"4","numAddedChangeFiles":"1","executionTimeMs":"2611","numDeletedRows":"1","scanTimeMs":"2236","numAddedFiles":"1","rewriteTimeMs":"374"},"engineInfo":"Apache-Spark/3.3.0 Delta-Lake/2.1.0","txnId":"ba662482-47fc-48f8-b338-9c6eafc46c39"}}
Reading 1 - actions from the commit log
In the reading part Delta Lake gets all changes from the commit log, reads them, and creates the final DataFrame. Sounds simplistic, doesn't it? Indeed, the process is a bit more complex than that. All starts in the DeltaSourceCDCSupport with the CDC files fetching:
trait DeltaSourceCDCSupport { self: DeltaSource =>
// ...
protected def getCDCFileChangesAndCreateDataFrame(
startVersion: Long,
startIndex: Long,
isStartingVersion: Boolean,
endOffset: DeltaSourceOffset): DataFrame = {
val changes: Iterator[(Long, Iterator[IndexedFile])] =
getFileChangesForCDC(startVersion, startIndex, isStartingVersion, None, Some(endOffset))
val groupedFileActions: Iterator[(Long, Seq[FileAction])] =
changes.map { case (v, indexFiles) =>
(v, indexFiles.map { _.getFileAction }.toSeq)
}
// ...
In the highlighted part, Delta Lake calls a private method that analyze the content of each commit file from the _commit_log and takes only the CDC actions, such as AddFile and RemoveFile if their dataChange attribute is true, or AddCDCFile that represents a CDC event file written to the _change_data directory. When should you expect each case?
- INSERT statement or DELETE operation without condition → the commit log should contain only AddFile and RemoveFile actions.
- Any other operation → should generate at least one AddCDCFile with all change events to return to the CDC consumer.
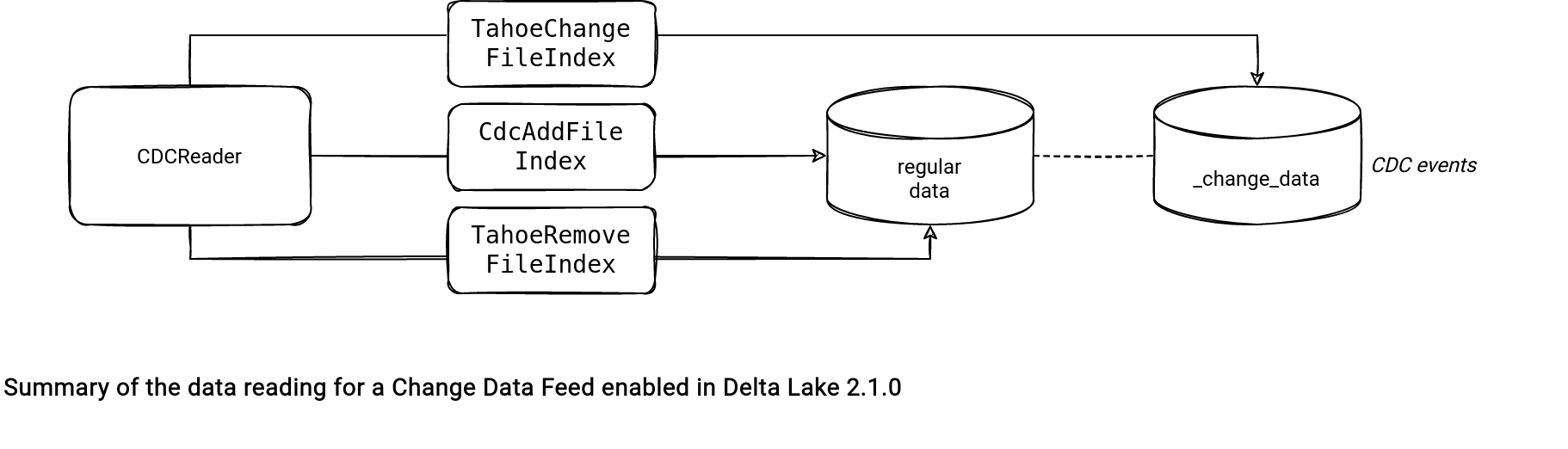
Reading 2 - indexes in the CDCReader
The next class involved in the reading is CDCReader. It takes the list of filtered action files and generates a DataFrame in the changesToDF( deltaLog: DeltaLog, start: Long, end: Long, changes: Iterator[(Long, Seq[Action])], spark: SparkSession, isStreaming: Boolean = false). The data generation logic uses TahoeFileIndex adapted to the action files:
- Deletes use a TahoeRemoveFileIndex.
- Inserts use a CdcAddFileIndex.
- Data changes use a TahoeChangeFileIndex.
The main difference between them is the matchingFiles(partitionFilters: Seq[Expression], dataFilters: Seq[Expression]): Seq[AddFile] that returns all valid files to the query.:
- TahoeChangeFileIndex converts the input Seq[CDCDataSpec[AddCDCFile]] to a list of AddFiles expected by the method signature.
- TahoeRemoveFileIndex also converts the input to the AddFiles but also adds one important attribute which is the _change_type=delete. I bet you can see it already. Indeed, thanks to this operation, the reader knows that the whole content of this file should be streamed as delete event!
- CdcAddFileIndex is quite similar to the TahoeRemoveFileIndex but instead of the "delete" action, it annotates the returned AddFile with the _change_type=insert.
The CDC in Delta Lake is yet another trade-off you can meet in the modern Table file formats between a fast write with slow read vs. a slow write with a fast read. The writing operation involves a few extra steps and IO with the extra files generated. On the other hand, the reading is fast because the write overhead has prepared all files for an efficient reading.
Consulting

With nearly 17 years of experience, including 9 as data engineer, I offer expert consulting to design and optimize scalable data solutions.
As an O’Reilly author, Data+AI Summit speaker, and blogger, I bring cutting-edge insights to modernize infrastructure, build robust pipelines, and
drive data-driven decision-making. Let's transform your data challenges into opportunities—reach out to elevate your data engineering game today!
👉 contact@waitingforcode.com
🔗 past projects


