The Scala API of Apache Spark SQL has various ways of transforming the data, from the native and User-Defined Function column-based functions, to more custom and row-level map functions. PySpark doesn't have this mapping feature but does have the User-Defined Functions with an optimized version called vectorized UDF!
Data Engineering Design Patterns

Looking for a book that defines and solves most common data engineering problems? I wrote
one on that topic! You can read it online
on the O'Reilly platform,
or get a print copy on Amazon.
I also help solve your data engineering problems 👉 contact@waitingforcode.com 📩
UDFs
Classical User-Defined Functions in PySpark are just like their Scala version, they work at the column basis. But unlike their Scala friends, they introduce a serialization/deserialization overhead which makes them difficult to use with other Python libraries like Pandas. As a consequence, the following function annotated with the @udf will be executed as many times as the number of rows in the input (20 times in my example):
numbers = spark.range(0, 20, numPartitions=5)
@udf(IntegerType())
def multiply(number: int) -> int:
print(number)
return number * 2
numbers_with_results = numbers.withColumn("result", multiply(col("id")))
numbers_with_results.show(truncate=False)
Is there a better way? Yes, starting from PySpark 2.3.0 you can use Vectorized UDFs!
Vectorized UDFs
A Vectorized UDF works on a subset of rows instead of one row at a time. It's then a big performance improvement compared to the classical UDF. Additionally it integrates with other popular Python tools, like Pandas. There is more! The definition of a Vectorized UDF consists of calling or decorating a Python function with...pandas_udf!
@pandas_udf(IntegerType())
def multiply(numbers_vector: pandas.Series) -> pandas.Series:
print(numbers_vector)
return numbers_vector.apply(lambda nr: nr * 2)
I'm sure you also see a second difference. Indeed, the parameter is not a single value but a bunch of values exposed as a Pandas Series object. That's all for the user-facing behavior. However, there are few other things to explore.
Vectorized UDF is an UDF
Not only from the conceptual point of view but also in the code. Both standard and vectorized versions calls this method:
def _create_udf(
f: Callable[..., Any],
returnType: "DataTypeOrString",
evalType: int,
name: Optional[str] = None,
deterministic: bool = True,
) -> "UserDefinedFunctionLike":
# Set the name of the UserDefinedFunction object to be the name of function f
udf_obj = UserDefinedFunction(
f, returnType=returnType, name=name, evalType=evalType, deterministic=deterministic
)
return udf_obj._wrapped()
So how does PySpark distinguish a vectorized function from a standard one? The answer is hidden in the evalType of which can be:
- SQL_BATCHED_UDF. It's the type reserved for the standard UDF. It's the single one supported for the not vectorized version.
- SQL_SCALAR_PANDAS_UDF. It's the first type for the Vectorized UDFs.
- SQL_SCALAR_PANDAS_ITER_UDF. It's similar to the scalar type presented above except that it takes an iterator of batches in the input instead of a single batch. It's useful if the UDF must initialize some shared state. The signature of the functions eligible for this type are Iterator[Tuple[Series, Frame or Union[DataFrame, Series], ...] -> Iterator[Series or Frame] and Iterator[Series, Frame or Union[DataFrame, Series]] -> Iterator[Series or Frame]
- SQL_GROUPED_MAP_PANDAS_UDF. The type is used by the FlatMapGroupsInPandasExec physical node that represents the UDF for the (pandas.DataFrame) -> pandas.DataFrame signature.
- SQL_GROUPED_AGG_PANDAS_UDF. Used for the aggregations expressed as UDFs, including the Series, Frame or Union[DataFrame, Series], ... -> Any Vectorized UDF signature .
- SQL_WINDOW_AGG_PANDAS_UDF. This one exists for windowed aggregates.
- SQL_MAP_PANDAS_ITER_UDF. It's the evalType defined in the function called by the mapInPandas.
- SQL_COGROUPED_MAP_PANDAS_UDF. It's the type used internally inside the grouping function passed to the applyInPandas.
- SQL_MAP_ARROW_ITER_UDF. PySpark uses this one for the custom function passed to the mapInArrow method.
PySpark passes this type to the Scala object representing the UDF and it's the Scala Spark responsibility to get the matching data.
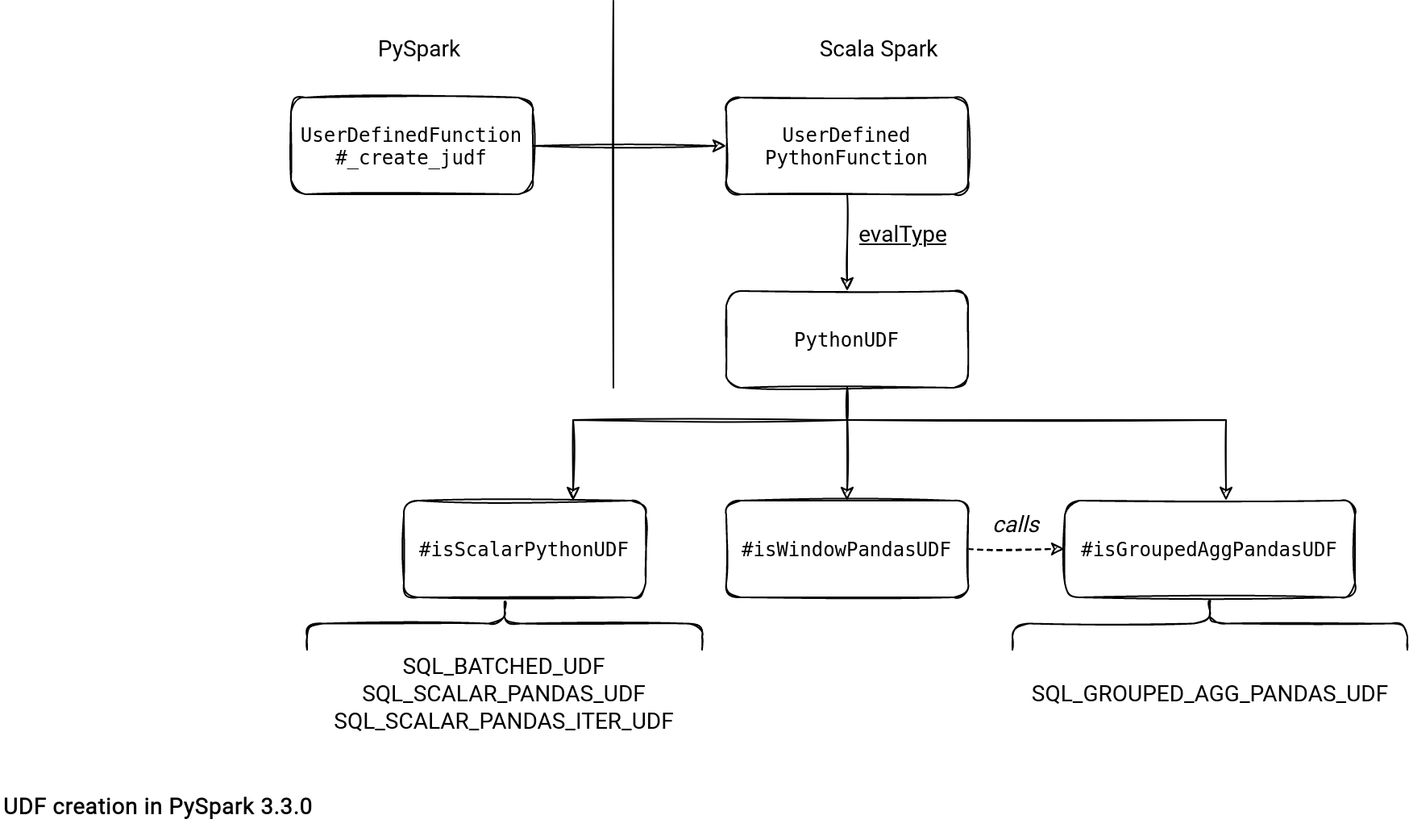
The evalType is not only useful to detect the type of the function on the Scala Spark side. It's also useful to transform the logical plan, like below for a Vectorized UDF from my previous code snippet:
== Analyzed Logical Plan == id: bigint, result: int Project [id#0L, multiply(id#0L)#2 AS result#3] +- Range (0, 20, step=1, splits=Some(5)) == Optimized Logical Plan == Project [id#0L, pythonUDF0#16 AS result#3] +- ArrowEvalPython [multiply(id#0L)#2], [pythonUDF0#16], 200 +- Range (0, 20, step=1, splits=Some(5))
How? Scala Spark has a logical rule called ExtractPythonUDFs that replaces the UDF part with ArrowEvalPython or BatchEvalPython. The former is the logical node representing a Vectorized UDF whereas the later applies to the standard function:
object ExtractPythonUDFFromAggregate extends Rule[LogicalPlan] {
// ...
private def extract(plan: LogicalPlan): LogicalPlan = {
// ...
val evaluation = evalType match {
case PythonEvalType.SQL_BATCHED_UDF =>
BatchEvalPython(validUdfs, resultAttrs, child)
case PythonEvalType.SQL_SCALAR_PANDAS_UDF | PythonEvalType.SQL_SCALAR_PANDAS_ITER_UDF =>
ArrowEvalPython(validUdfs, resultAttrs, child, evalType)
case _ =>
throw new IllegalStateException("Unexpected UDF evalType")
}
What's the difference between them? The BatchEvalPython transforms into BatchEvalPythonExec physical node that communicates with PySpark at the row level. On the other hand, the ArrowEvalPython gets handled by the ArrowEvalPythonExec that exchanges the data via an Apache Arrow stream. The number of records exchanged at once depends on the value set in the spark.sql.execution.arrow.maxRecordsPerBatch which by default is 10000.
On the PySpark side, worker.py exposes a read_udfs(pickleSer, infile, eval_type) function that deserializes data from the JVM with a serializer corresponding to the evalType:
# worker.py
def read_udfs(pickleSer, infile, eval_type):
# ...
if eval_type == PythonEvalType.SQL_COGROUPED_MAP_PANDAS_UDF:
ser = CogroupUDFSerializer(timezone, safecheck, assign_cols_by_name)
elif eval_type == PythonEvalType.SQL_MAP_ARROW_ITER_UDF:
ser = ArrowStreamUDFSerializer()
else:
# Scalar Pandas UDF handles struct type arguments as pandas DataFrames instead of
# pandas Series. See SPARK-27240.
df_for_struct = (
eval_type == PythonEvalType.SQL_SCALAR_PANDAS_UDF
or eval_type == PythonEvalType.SQL_SCALAR_PANDAS_ITER_UDF
or eval_type == PythonEvalType.SQL_MAP_PANDAS_ITER_UDF
)
ser = ArrowStreamPandasUDFSerializer(
timezone, safecheck, assign_cols_by_name, df_for_struct
)
else:
ser = BatchedSerializer(CPickleSerializer(), 100)
Vectorized UDF integrates better with Python world and also relies on Apache Arrow. After writing this yet another PySpark blog post I feel that it's a missing piece in my exploration. That's why you can expect some Apache Arrow-related articles in the coming weeks!
Consulting

With nearly 17 years of experience, including 9 as data engineer, I offer expert consulting to design and optimize scalable data solutions.
As an O’Reilly author, Data+AI Summit speaker, and blogger, I bring cutting-edge insights to modernize infrastructure, build robust pipelines, and
drive data-driven decision-making. Let's transform your data challenges into opportunities—reach out to elevate your data engineering game today!
👉 contact@waitingforcode.com
🔗 past projects


